5-substituted uracil derivatives as the potential radiosensitizers
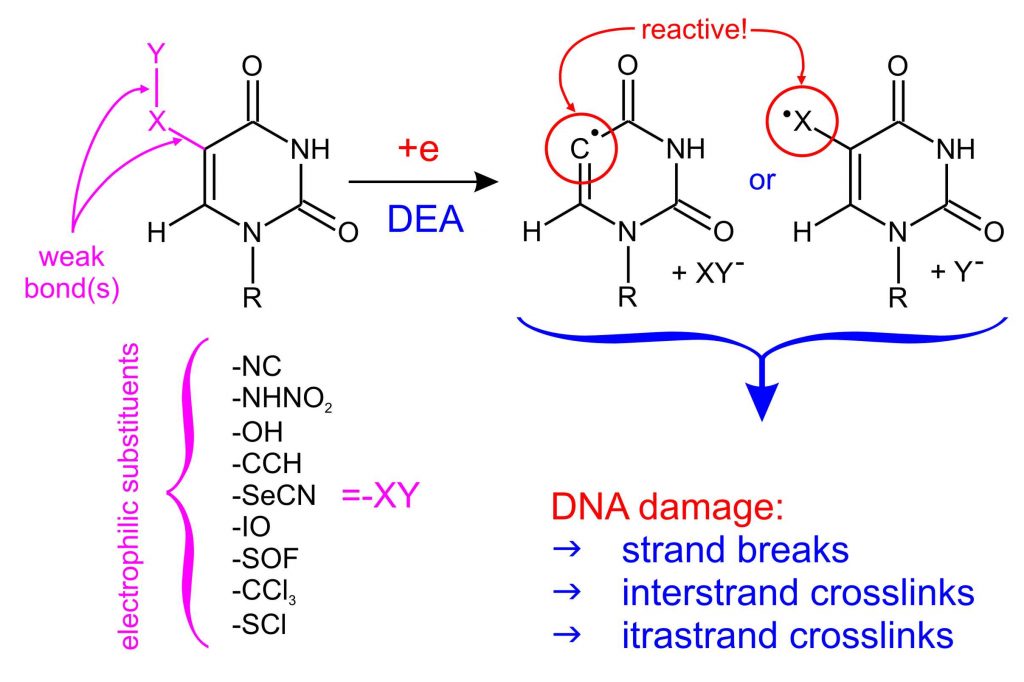
To pre-select potential radiosensitizers from the proposed MNs, we developed a simple and low-cost protocol based on quantum chemical (DFT) calculations that analyzes the behavior of MNs during the attachment of an electron (ChemPhysChem, 2016, 17, 1-8). The reactive radical formation in a nucleobase (or a substituent’s fragment) in a quantum-chemically modeled DEA reaction is a premise for further research on the sensitizing properties of the proposed compound (Figure 1). The proposed basic MNs pre-selection protocol, on the one hand, allowed to reject dozens of uracil derivatives, which, after electron attachment, create stable anion radicals incapable of DEA process, without the necessity of costly synthesis and analysis of not promising compounds. On the other hand, it has already allowed to propose several promising uracil derivatives, potential radiosensitizers: SeCNdU, OTfdU, BrSdU, ISdU, CF3CONHU, SU, DMSU or 6IdU.
Modeling of damage in double-stranded DNA labeled with sensitizing MNs.
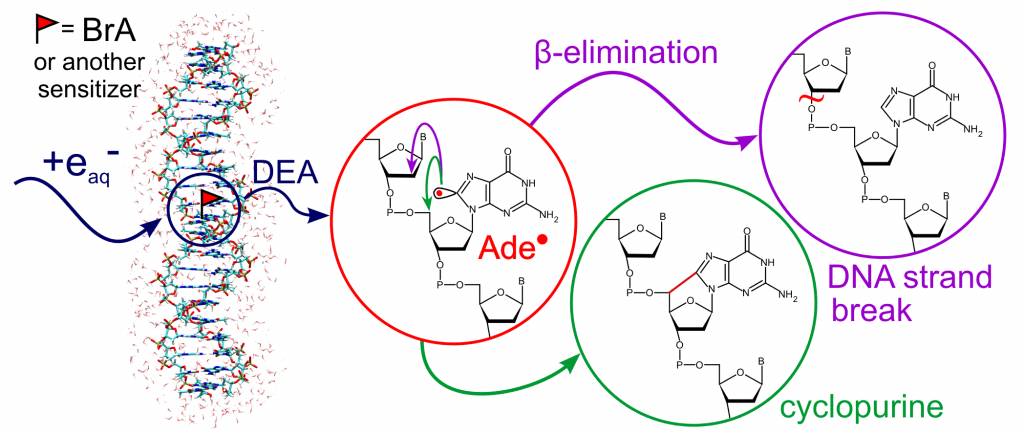
Obtaining a reactive radical inside the DNA strand as a result of the degradation of the potential radiosensitizer is considered to be crucial to initiate a series of radical reactions, ultimately leading to permanent damage of cell DNA. Using the QM/MM metadynamics, we comprehensively analyzed the possible degradation pathways of the radical formed from MN, (J. Chem. Theory Comput. 2017, 13, 6415–6423). The applied methodology allowed to take into account the DNA double-strand environment and, for the first time, to fully describe the thermodynamics and kinetics of the experimentally observed purine derivatives degradation paths : β-elimination (responsible for the formation of strand breaks) and cyclization (Figure 2).
Stationary and pulsed radiolysis of aqueous solutions containing potential radiosensitizers.
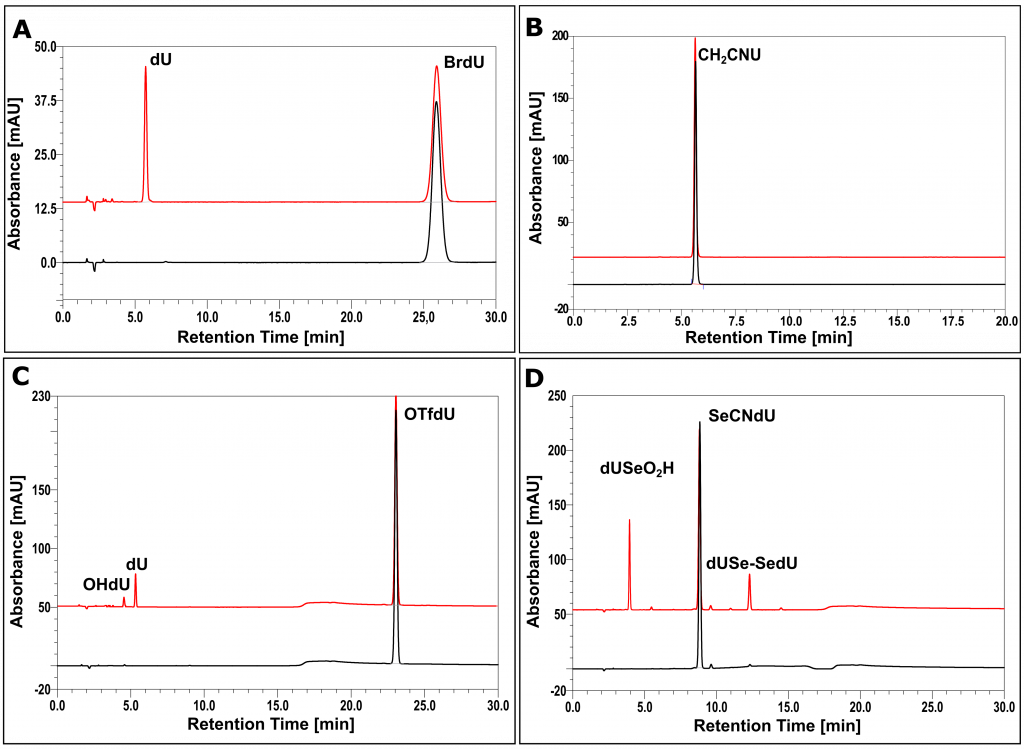
One of the necessary conditions for MN to become a radiosensitizer is its degradation induced by the electron attachment. The stationary radiolysis experiments of deoxygenated, aqueous solutions of tested derivatives in the presence of phosphate buffer and tert-BuOH (scavenger of free radical •OH) allowed for preliminary verification of radiosensitizing properties of individual derivatives (Figure 3). In the case of SeCNdU, OTfdU (RSC Adv., 2018, 8, 21378−21388), ISdU (IJMS, 2019, 20, 1308), BrSdU (Molecules, 2019, 24, 2819) and CF3CONHU (IJMS, 2020, 21, 6352), the degradation products were identified and their structures were determined using the LC-MS/MS technique. The mechanisms of radioproducts formation were also proposed and confirmed by pulse radiolysis experiments (Molecules, 2019, 24, 2819).
Studies at the cellular level of promising systems.
The derivatives that passed the initial verification of their radiosensitizing properties were tested at the cellular level. MTT/WST-1 tests were performed to determine cytotoxicity against normal (HDFa) and cancer cells (MCF-7 and PC3). The radiosensitizing properties of MNs against cancer cells were confirmed on the basis of a clonogenic assay.
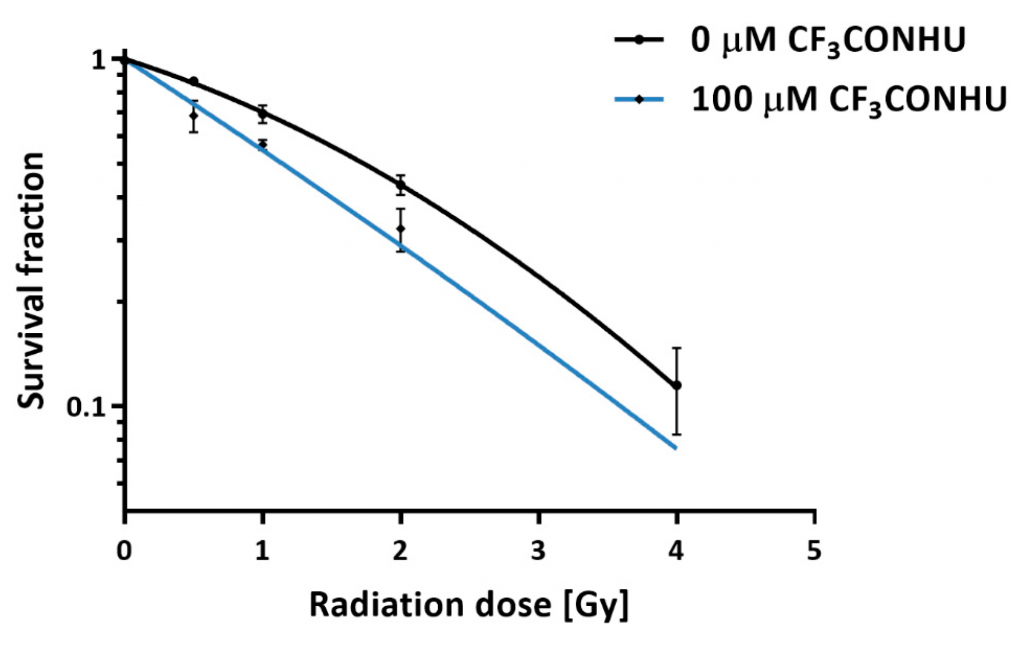
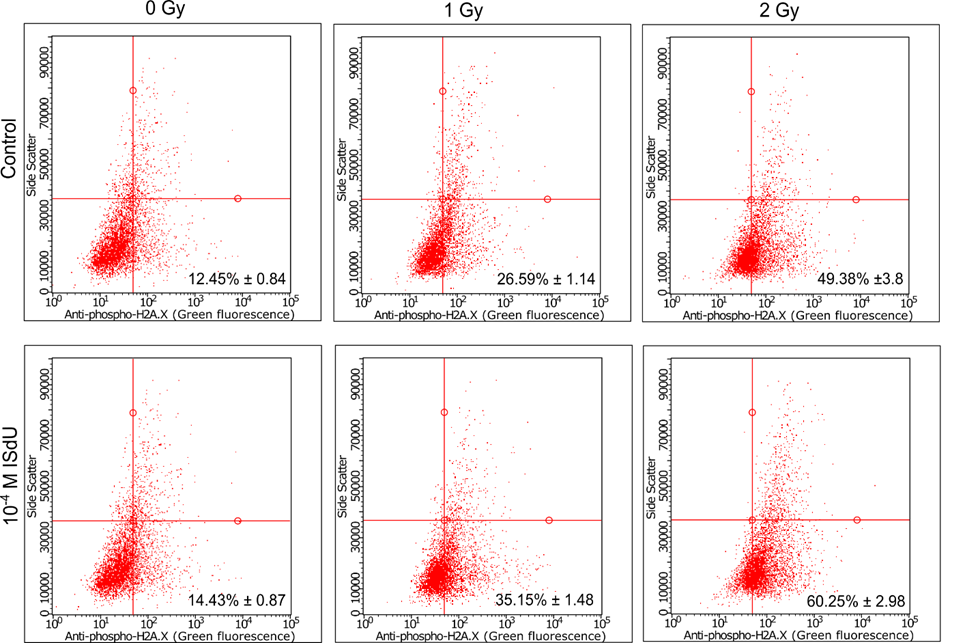
In the case of cells incubated with ISdU (IJMS, 2019, 20, 1308) and CF3CONHU (IJMS, 2020, 21, 6352) (Figure 4), a decrease in the survival rates was observed. In the case of BrSdU (Molecules, 2019, 24, 2819), no differences in survival rates were observed for the cells with or without the derivative. This fact confirms once again the key role of the DEA process in radiosensitization. In the case of ISdU and BrSdU, additional cellular tests were performed. Cytometric analysis of the DNA double strand breaks’ marker – histone H2A.X – indicates that treatment of cells with ISdU (IJMS, 2019, 20, 1308) increases the pool of γH2A.X-positive cells (Figure 5), and confirms, that DNA double strand breaks are responsible for the increased sensitivity of cells to ionizing radiation (IR).
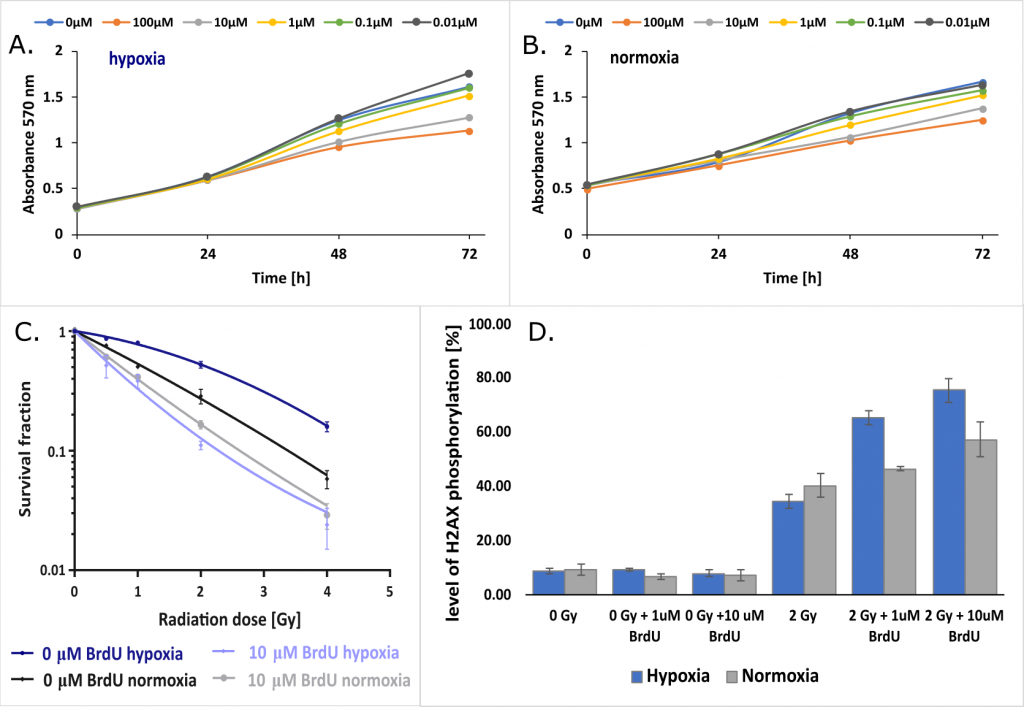
Another project investigated the effect of hypoxia on the radiosensitizing properties of BrdU. It turned out that while the oxygen concentration did not significantly affect for the efficiency of nucleoside incorporation into genomic DNA and the proliferation of tumor cells from the MCF-7 and PC3 lines (Figures 6A and 6B), the radiosensitizing effect clearly depends on the degree of their oxygenation (Figure 6C). Further studies on the mechanism of radiosensitization at the cellular level showed that the formation of DNA double-strand breaks induced by IR (histone H2A.X – the molecular marker of phosphorylation) is definitely more efficient under hypoxic conditions than under normal oxygen concentration (Figure 6D). In addition, the major checkpoints of the cellular response to DNA damage induced by IR were analyzed with western blot. “Influence of hypoxia on radiosensitization of cancer cells by 5-bromo-2’-deoxyuridine” by M. Zdrowowicz, P. Spisz, A. Hać, A. Herman-Antosiewicz, J. Rak is currently being published.
Enzymatic synthesis of labeled DNA.
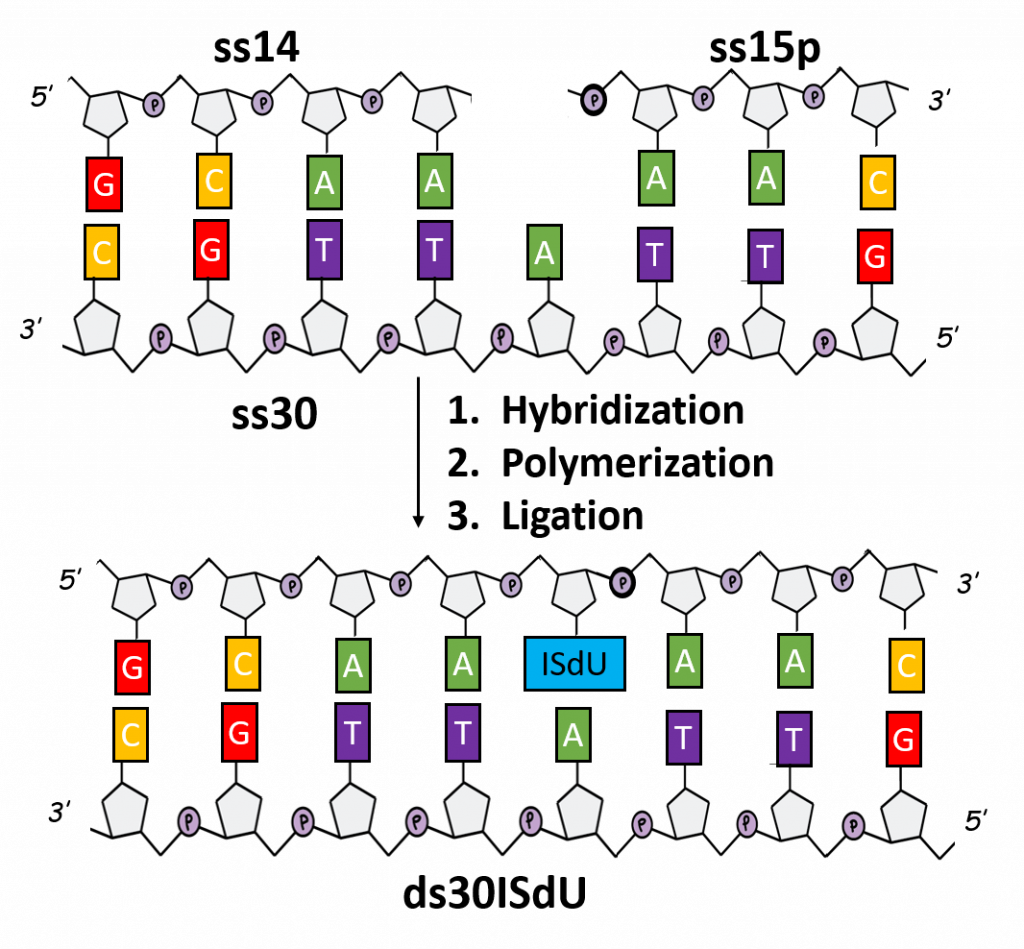
The search of an effective sensitizer is inherently connected with the use of labeled DNA sequences. However, the synthesis of such systems is severely limited due to the availability of the corresponding triphosphates/amidophosphates. As part of this project, the chemical synthesis of 5-iodo-4-thio-2′-deoxyuridine-5′-triphosphate (ISdUTP) and the enzymatic synthesis of DNA fragment model (30 bp), marked with ISdU (Figure 7B).
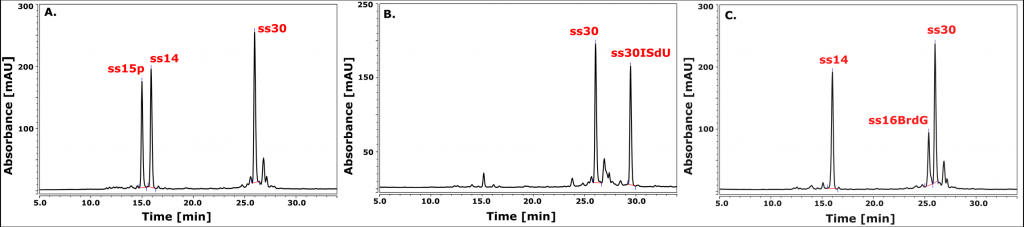
The enzymatic synthesis constituted of successive hybridization of three complementary oligonucleosides (ss14, ss15p and ss30), incorporation of ISdUTP by an isothermal polymerase (exo- Klenow fragment) and joining the resulting ends by T4 ligase (Figure 8). A similar approach was used for the synthesis of the BrdG-labeled DNA fragment (Figure 7C). In this case, optimization of the ligation step is necessary. Optimized synthesis protocols will enable further experiments on the mechanistic details of the processes of both photo- and radiosensitization and on indicating the type of DNA damage. „Chemically-enzymatic synthesis and photosensitivity of DNA labeled with 5-iodo-4-thio-2′-deoxyuridine” by M. Zdrowowicz, A. Czaja, W. Kozak and J. Rak is waiting to be published.
Mechanism of photoinduced damage to labeled DNA.
The photosensitizing properties of BrdU have been studied since the 1960s, and the mechanism of sensitization caused by the presence of this halogen uracil derivative in DNA has been attributed to long-range electron transfer (ET) caused by electron excitation of BrdU by the UVB light (λmax ~ 280 nm). The research carried out under this project confirmed that hypothesis. More specifically, we investigated the efficiency of photochemical DNA single-strand break (SSB) formation in synthetic double-stranded oligonucleotide sequences, 5′-XGGABrUY, labeled with BrU and differing in X, Y bases adjacent respectively to the 3′ side of BrdU and the 5′ side of guanine. The efficiency of strand break formation was determined by the LC/MS method, and the mathematical model, describing the dependence of efficiency of the SSB formation on the susceptibility of the oligonucleotide to ET, was created and derived on Marcus’s theory of electron transfer:
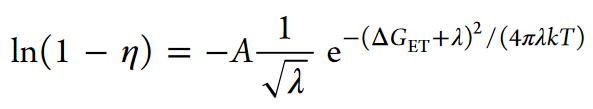
where η is the efficiency of SSB formation, ΔGET is the magnitude of the thermodynamic stimulus of the ET process, and λ is the reorganization energy. The thermodynamic stimulus, ΔGET, was calculated as the difference in electron affinity and ionization energies as calculated with QM, for each BrUY: XG pair.
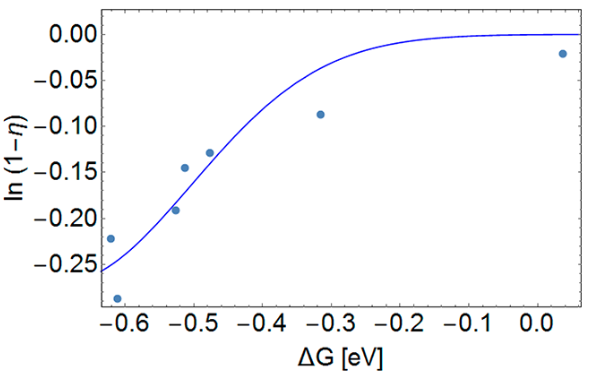
The fact that the model reflects the experimental points distribution (Figure 9) confirms photo-induced electron transfer as the primary cause of SSB formation in the tested systems (J. Phys. Chem. B, 2017, 121, 9169−9174).
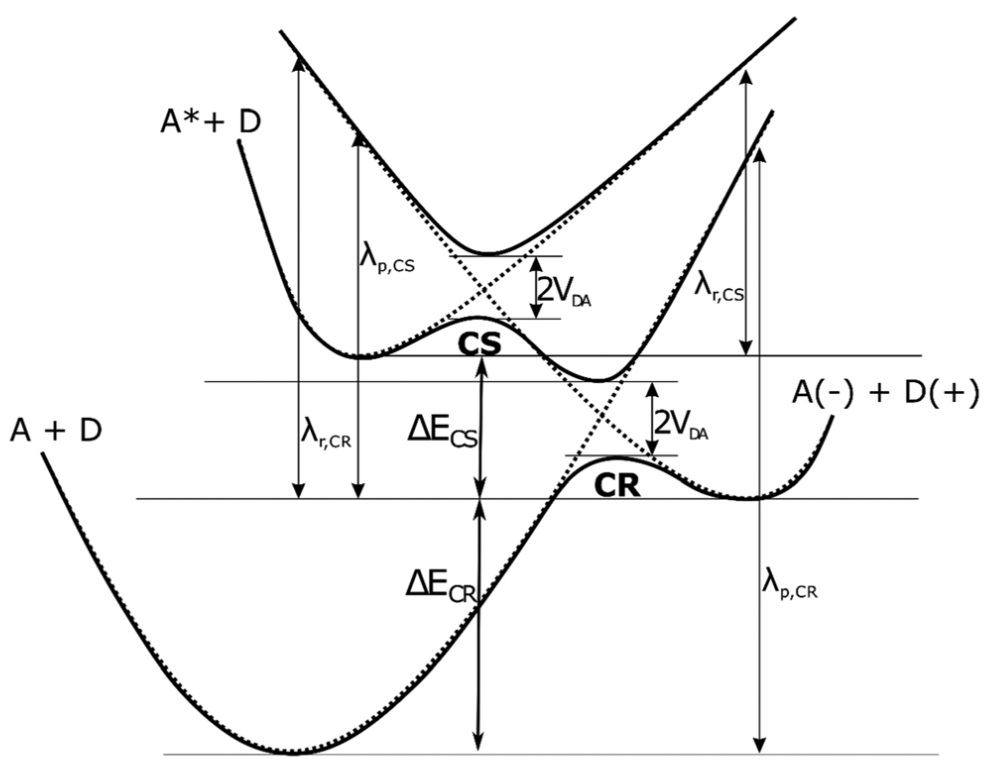
Similarly, our studies explaining the significantly higher efficiency of photoinduced SSB formation in the 5′-ABrU sequence compared to 5′-GBrU in double-stranded DNA, suggested a mechanism of damage through photoinduced ET from the distant guanine (Phys. Chem. Chem. Phys., 2019, 21, 4387−4393). Indeed, the electron coupling values determined by the FCD method (and averaged over 3000 configurations extracted from the MD trajectories for a double-stranded oligonucleotide with the sequence 5′-GCG CTA TCG XBrUT ATC GCG, where X = A or G), the reorganization energy and the driving force of the process (Figure 10), show that due to the relatively high energy of reorganization, the rate constants of the ET process are too low to lead to efficient photo-damage of DNA. Only the introduction of the (A)n bridge between the electron donor (G) and BrU leads (thanks to electron delocalization) to the reduction of the energy reorganization and a corresponding increase in the rate constant of electron transfer, which enables the progress of the photochemical SSB formation reaction. Our results perfectly explain the significant efficiency of photo-damage in the 5′-GAABrU sequence and the lack of them in the 5′-AAGBrU sequence.
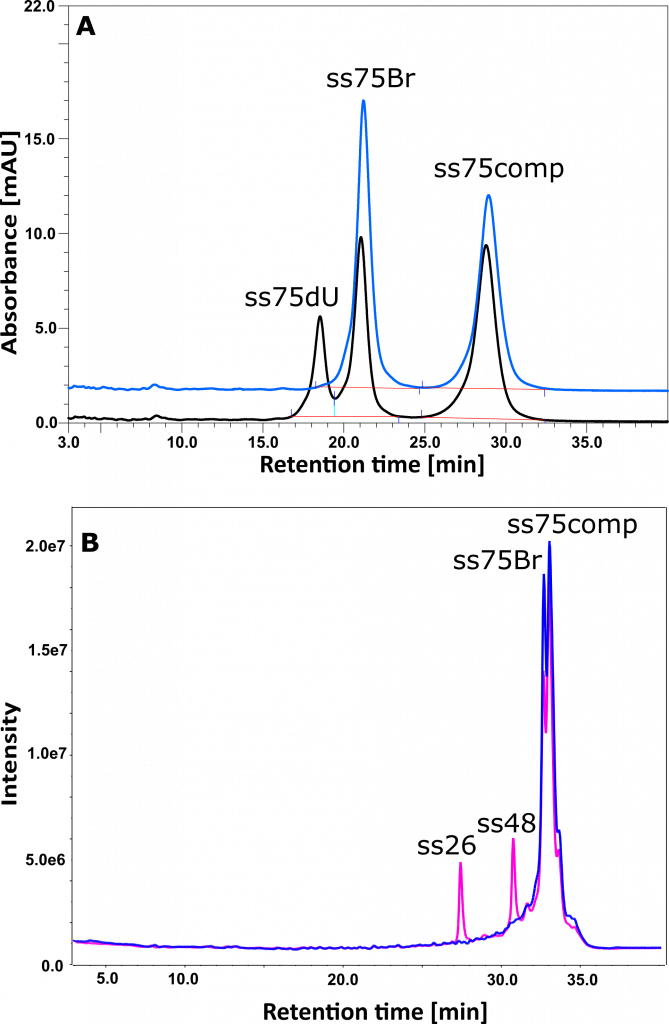
The photochemical research described above also led to a completely unexpected observation. Namely: the absence of breaks in the biopolymer in some photochemical experiments, while photodebromination of BrdU. An example of such experiment is shown in Figure 11 (J. Pharm. Biomed. Anal., 2017, 142, 262–269). The 11A chromatogram shows the photochemical formation of an oligonucleotide containing 2′-deoxyuridine instead of BrdU. In contrast, the 11B chromatogram shows the formation of break in the same sequence (fragments ss26 and ss48). The difference between the irradiated systems comes down to the different methods of oligonucleotide preparation. The DNA fragment used in experiment (11A) comes directly from the manufacturer, while the DNA fragment used in experiment (11B) was repeatedly lyophilized before being dissolved in an appropriate buffer. We explained this effect by contamination of commercial oligonucleotides with triethylamine (TEA; IP-RP-HPLC chromatography is used to purify synthetic oligonucleotides, where triethylamine acetate is used as a factor enabling the formation of ion pairs). Indeed, TEA is a known electron donor leading to the efficient formation of ion pairs in photochemical reactions (e.g. excitation of cyclohexanone in the presence of TEA). This charge transfer reaction is probably responsible for the complete extinction of the long-range ET inside the DNA, which, in the presence of TEA, is replaced by direct electron transfer from triethylamine to BrdU, leading to the formation of dU. Using mass spectrometry, we have shown that, depending on the producer (materials from 4 producers were tested), the amount of triethylamine exceeds even 1000 times the amount of oligonucleotide in the ordered material.
Enzymatic phosphorylation of MN – a necessary condition of the sensitizer’s function.
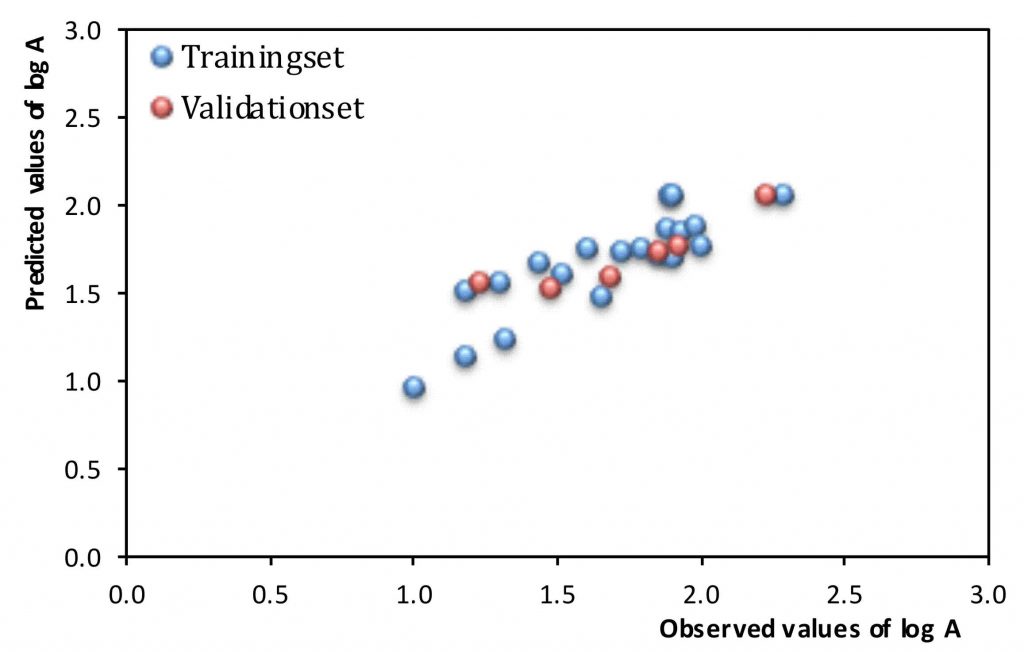
In order to work effectively, nucleosides must incorporate into DNA during its replication or repair. One of the basic conditions for incorporation into DNA is fitting nucleoside to the hTK1 substrate’s specificity, which is an enzyme that catalyzes the conversion of nucleosides into their 5′-monophosphates. In Struct. Chem. 2018, 29, 1367–1374, we published a chemometric model predicting hTK1 activity towards nucleosides (Figure 12). For the construction of the model, we used the enzymatic activity of hTK1, determined experimentally for 26 uridine analogues. We obtained a reliable QSAR model requiring only three molecular descriptors. Two of them relate to the shape of the molecule, and the third is related to its susceptibility to interactions with the enzyme. The conducted research also suggested that the most easily phosphorylated should be 5-substituted uridine analogs.
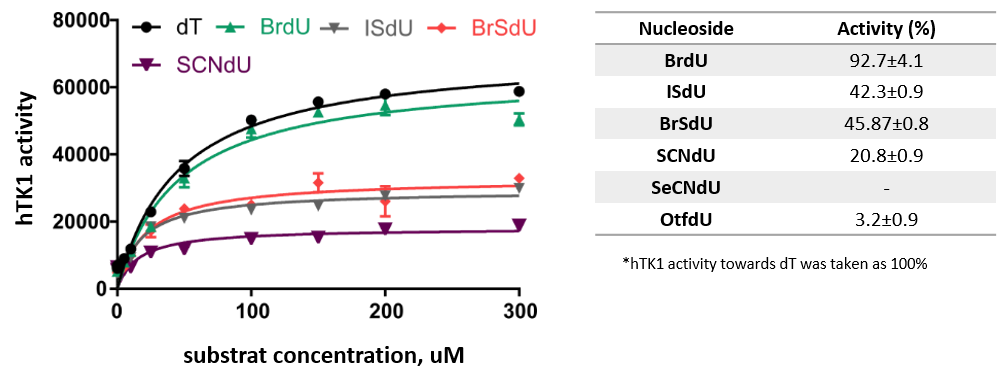
The hTK1 kinase activity against modified nucleosides with sensitizing properties was also determined experimentally. Kinetic studies were consisted in monitoring the emerging ADP by spectrofluorometry. Figure 13 shows the activity of the kinase against the tested modified nucleosides. A significant decrease in activity was observed in the case of the OTfdU derivative, while SeCNdU occurred to be not phosphorylated at all in the tested system. The obtained results may suggest that intracellular phosphorylation is a „bottleneck” of the incorporation of these analogs into genomic DNA.
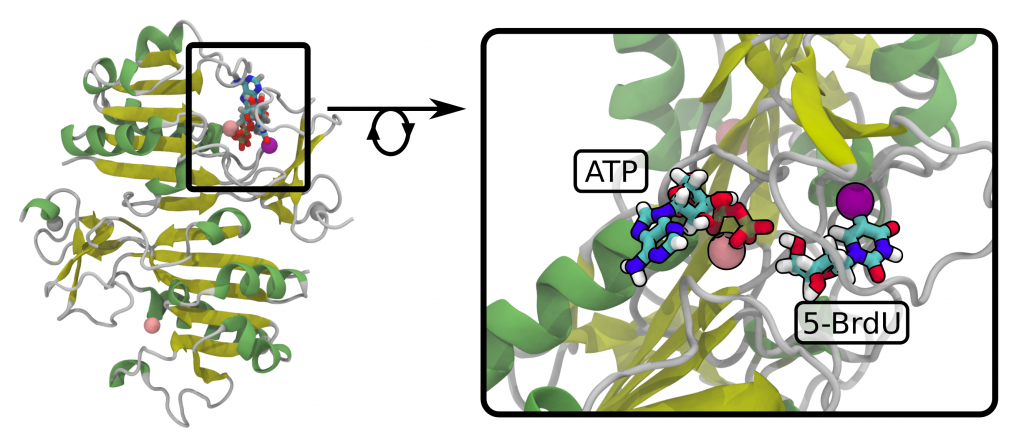
The effectiveness of hTK1 should correlate with the nucleoside’s binding energy to the active site. Differences in the affinity of hTK1 thymidine kinase for modified nucleosides, with respect to its native substrate (thymidine, dT), were estimated using an „alchemical” variant of molecular dynamics. In this methodology dT is gradually transformed into the test ligand both in the active site of the enzyme and in the solution, and the free energy changes of these transformations are determined on the basis of the thermodynamic integration method. Subtracting the above-mentioned changes in free energy allows, using an appropriate thermodynamic cycle, to calculate the values of ΔΔG of the interaction with respect to the interaction with dT, characterizing the ability of the ligands to binding to hTK1 (the values of ΔΔG for the tested nucleosides: BrdU, SCNdU, OCNdU, SeCNdU and OTfdU are -1.76, +1.79, -3.03, +2.44 and +0.52 kcal/mol respectively). An additional effect of simulations was obtaining the optimized structures of modified nucleosides in the hTK1 active center (Figure 14).
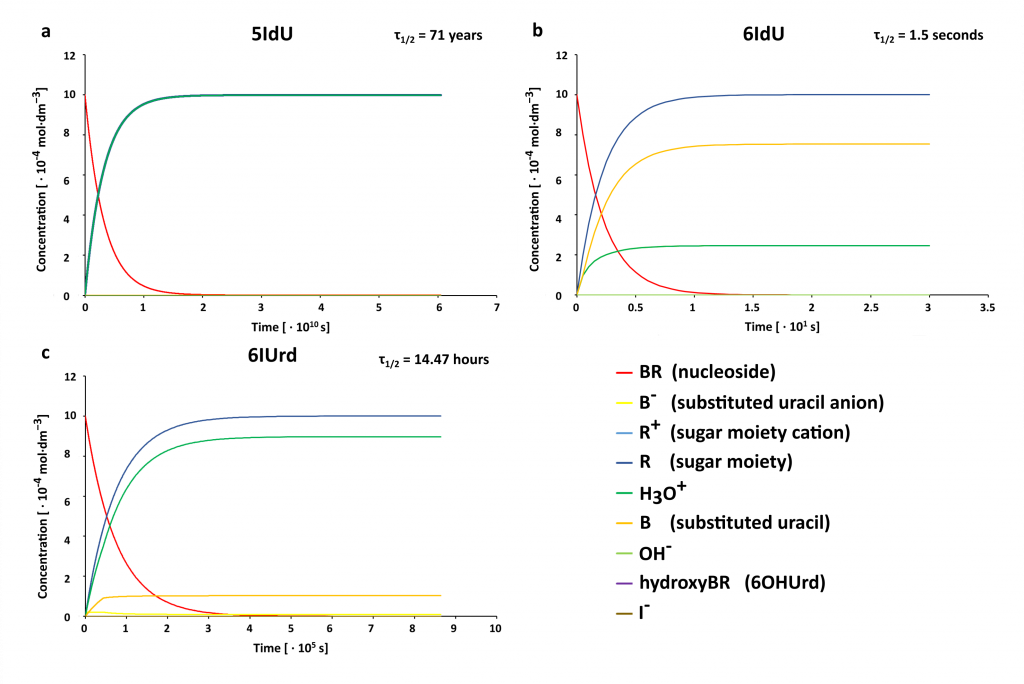
Stability of MN in water solution.
Since water is the natural environment of all biological systems, modified nucleosides must be stable in that environment to effectively label DNA. We characterized the process of hydrolysis of selected uracil derivatives using the DFT method, which was used to determine the activation energy and thermodynamic stimulus for individual elementary reactions. The kinetic barrier for the 6-iodouridine hydrolysis reaction (6IUrd) was also determined experimentally by constructing the Arrhenius plot and was used to verify the adopted level of theory (CAM-B3LYP/DGDZVP++). The theoretically calculated characteristics of individual reactions were used to construct a system of differential equations, which results are presented in Figure 15. The striking differences of the half-time for the tested derivatives clearly show that 6IdU cannot act as a radiosensitizer (τ1/2 is only 1.5 seconds – Figure 15). Therefore, prediction of the hydrolytic stability of modified nucleosides should precede their synthesis. The work on this topic will soon be sent to the JPC B.